
Oxford Nanopore has developed a new generation of DNA/RNA sequencing technology. It is the only sequencing technology that offers real-time analysis (for rapid insights), in fully scalable formats from pocket to population scale, that can analyse native DNA or RNA and sequence any length of fragment to achieve short to ultra-long read lengths.
Nanopores
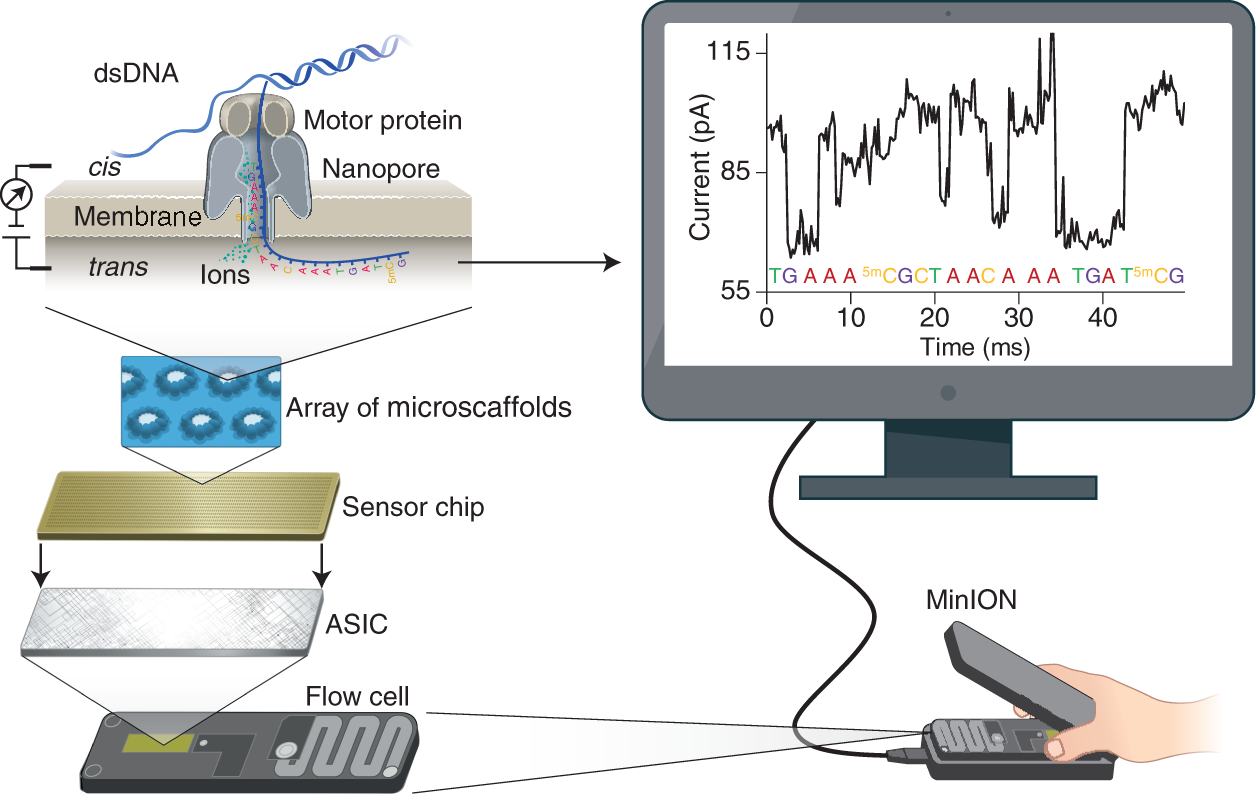
All Oxford Nanopore sequencing devices use flow cells which contain an array of tiny holes — nanopores — embedded in an electro-resistant membrane. Each nanopore corresponds to its own electrode connected to a channel and sensor chip, which measures the electric current that flows through the nanopore. When a molecule passes through a nanopore, the current is disrupted to produce a characteristic ‘squiggle’. The squiggle is then decoded using basecalling algorithms to determine the DNA or RNA sequence in real time.
Learn more about flow cells and nanopores
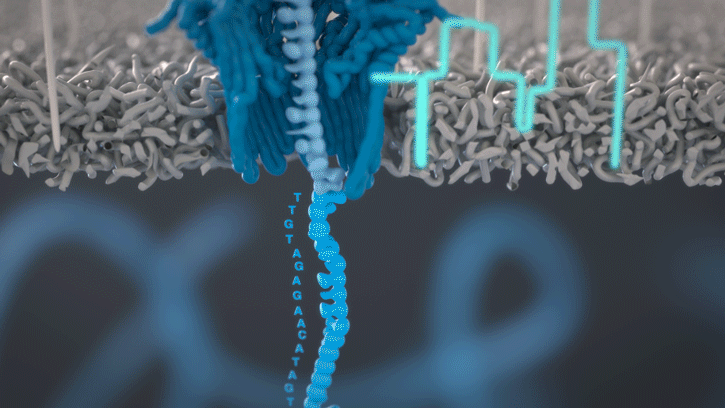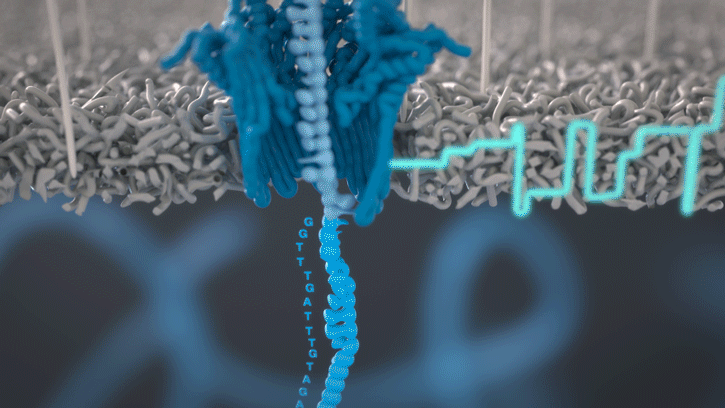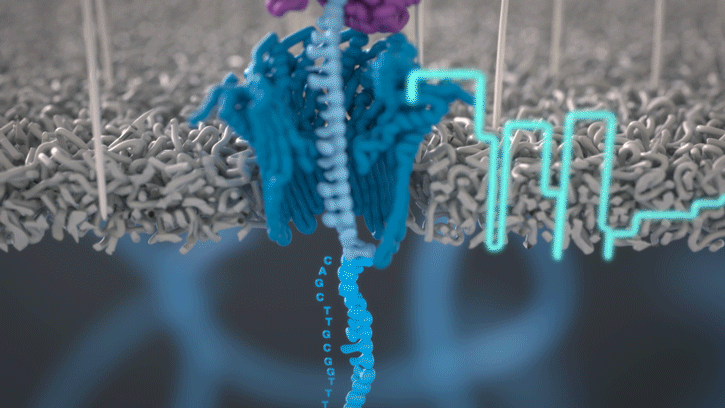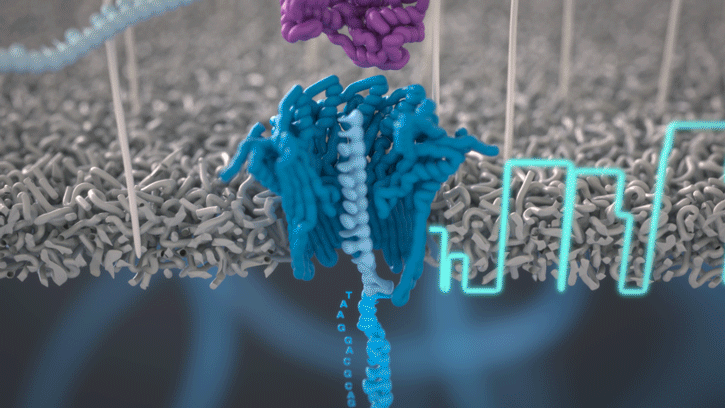
You can think of the current as water flowing through a pipe. When an object enters the pipe, the flow of water is disrupted, just as DNA disrupts the current as it passes through the nanopore.

DNA/RNA sequencing
A strand of DNA or RNA is made up of a sequence of different combinations of four nucleotide bases: A, T (or U for RNA), G and C. Each base that passes through the nanopore can be identified through the characteristic disruption it causes to the current in real-time. This makes nanopore sequencing unique, in that it is the only sequencing technology that enables direct, real-time analysis of short to ultra-long fragments of DNA/RNA, in fully scalable formats. Advantages of real-time sequencing include rapid access to time critical information (e.g. pathogen identification), the generation of early sample insights and more control over the sequencing experiment.















